Molecular detection of virulence genes in Klebsiella Pneumoniae clinical isolates from Kurdistan Province, Iran
- Cellular and Molecular Research Center, Kurdistan University of Medical Sciences, Sanandaj, Iran
- Department of Microbiology, School of Medicine, Kurdistan University of Medical Sciences, Sanandaj, Iran
- Social Determinants of Health Research Center, Lorestan University of Medical Sciences, Khorramabad, Iran
- Iran National Science Foundation (INSF)
Abstract
Introduction: The purpose of this study was to detect ybtS, entB, mrkD, magA, kfu, iutA, rmpA and K2 genes in extended-spectrum beta-lactamase (ESBL) - and non-ESBL producing Klebsiella pneumoniae.
Methods: To this end, 70 K. pneumoniae isolates were selected from hospitals of Kurdistan Province, Iran. The ESBL phenotype was conducted utilizing the disc diffusion technique in accordance with CLSI procedures. Detection of virulence factor genes was performed by the PCR in the ESBL and non-ESBL isolates.
Results: Sixty-two (88.6%) isolates of K. pneumoniae were ESBL producers. Further, entB had the most frequency in all the isolates. There were no significant differences between ESBL production and the presence of ybt S, entB, mrkD, magA, kfu, iutA, rmpA and K2 genes and the presence of these genes and variables such as presence of sex, clinical specimen type, and hvKP phenotype among the ESBL and non-ESBL K. pneumoniae isolates.
Conclusion: In conclusion, in other studies, K. pneumoniae strains were separated from liver abscesses and the magA gene was frequently present; however, in our study, the K. pneumoniae strains were separated from various clinical specimens and the magA gene had low frequency.
Background
is a prominent opportunistic pathogen which causes upper respiratory tract infection, diarrhea, pneumonia, urinary tract infection (UTI), and septicemia 123. The prevalence of drug resistance in has increased, which is because of extended-spectrum beta-lactamase (ESBL) enzymes and appearance of multi-drug resistant (MDR) 45. In addition, possesses different virulence factors that contribute to its pathogenicity including lipopolysaccharide (LPS) O-side chain (endotoxin), capsular polysaccharide, adhesions and sidrophores 647. The LPS contains lipid A, core, and O-polysaccharide antigen 8. Capsule polysaccharide (CPS) is a major factor for virulence of and classified into 77 serological types (K) 9. Capsular layers engulf the surface of bacteria and prevent bacteria phagocytosis. and capsular antigens are the most important ones 10.
Genome of the capsule comprises gene clusters (capsular polysaccharide synthesis), (mucoviscosity associated gene A), and wb (O-specific polysaccharide directed by the wb gene cluster) 11. (35-Kbp) was identified as a K1-specific capsular polymerase gene which acts as a trans-acting activator for biosynthesis of cps. Moreover, is homologous with the genes involved in glycosylation, transfer and biosynthesis of the LPS12. In 2004, was determined as the major virulence factor of 2. It has been reported that can magnify the colony mucoidy of different serotypes of and act as a plasmid-mediated regulator of extra capsular polysaccharide synthesis 13. Adhesives include Pilli, the building of protein, and the attachment of bacteria to the host. gene mediates binding to the extracellular matrix, and also codes type 3 fimbria adhesion 14. by different siderophores (iron-bound) including enterobactin, yersiniabactin and hydroxamate siderophore obtain iron from transferrin and lactoferrin in host transport proteins. and genes encode enterobactin, yersiniabactin, iron-uptake system and hydroxamate siderophore 15. The main purpose of the current study was to detect , and genes in ESBL and non-ESBL producing isolated from clinical specimens in Kurdistan Province, Iran.
Methods
Identification of bacterial strains
Seventy isolates were taken from specimens including urine, blood, tracheal aspirates and wound from October 2015 to July 2016 from general hospitals of Kurdistan Province, Iran. All the isolates were cultured on blood and MacConkey agar (Merck, Germany). Colonies were identified by Gram stain and biochemical tests such as urea hydrolysis, H2S production, lysine decarboxylase, lactose fermentation, indole, methyl red, voges proskauer, citrate (IMViC) and oxidase tests 16.
Phenotypic detection of ESBLs strains
Detection of ESBLs was tested by the combination disk diffusion test (CDDT) for the isolates. The CDDT was performed by ceftazidime (30 μg), cefotaxime (30 μg), cefepime (30 μg), cefpodoxime (30 μg), cefotaxime-clavulanic acid (30/10μg), cefotaxime-clavulanic acid (30/10 μg), cefepime-clavulanic acid (30/10 μg) and cefpodoxime-clavulanic acid (30/10 μg) (Roscoe, Denmark) (Ho 1998). ATCC 25922 and ATCC 7881 were utilized for negative and positive controls, respectively.
Determination of hypermucoviscosity K. pneumoniae (hv-KP) phenotype
Seventy isolates were separated from the clinical samples and cultivated on blood agar medium (Merck, Germany); then, they were incubated at 37°C for 24 h. Subsequently, the hypermucoviscosity of (hv-KP) phenotype was determined by forming a viscous string more than 5 in standard bacteriological loops 3.
Virulence genes identification by PCR amplification
The isolates were cultured on Luria broth (LB) medium overnight. Then, DNA samples were extracted using Genomic DNA Extraction Kit (SinaClon, Iran). Gene coding virulence factors were detected by the PCR method. PCRs were carried out by using the thermo cycler system (Bio-Rad, Australia) and master mix PCR (YT1553, Iran) and primers were designed by Compain (
Characteristics of the primers used in PCRs
| Primer | Name DNA sequence (5 to 3) | Amplicon size (bp) |
| ybtS_forybtS_rev | GACGGAAACAGCACGGTAAAGAGCATAATAAGGCGAAAGA | 242 |
| mrkD_formrkD_rev | AAGCTATCGCTGTACTTCCGGCAGGCGTTGGCGCTCAGATAGG | 340 |
| entB_forentB_rev | GTCAACTGGGCCTTTGAGCCGTCTATGGGCGTAAACGCCGGTGAT | 400 |
| rmpA_forrmpA_rev | CATAAGAGTATTGGTTGACAGCTTGCATGAGCCATCTTTCA | 461 |
| K2_forK2_rev | CAACCATGGTGGTCGATTAGTGGTAGCCATATCCCTTTGG | 531 |
| kfu_forkfu_rev | GGCCTTTGTCCAGAGCTACGGGGTCTGGCGCAGAGTATGC | 638 |
| iutA_foriutA_rev | GGGAAAGGCTTCTCTGCCATTTATTCGCCACCACGCTCTT | 920 |
| magA_formagA_rev | GGTGCTCTTTACATCATTGCGCAATGGCCATTTGCGTTAG | 1283 |
Statistical analysis
The association between the ESBL production, clinical specimen type, sex ,and hvKP phenotype and presence of genes among the ESBL and non-ESBL isolates was analyzed by Fisher tests with STATA software program v12.
Results
Bacterial isolates
Out of the 70 isolates, 37 (52.9%), 32 (45.7 %) and 1 isolates (1.4%) were collected from women, men and the hospital environment. Moreover, 50 isolates (71.4%) were obtained from urine, 8 isolates (11.4%) from blood, 10 isolates (14.3%) from tracheal aspirates, 1 isolate (1.4%) from wound, and 1 isolate (1.4%) from the environment (
The characteristics of the hypermucoviscosity clinical isolates
| Sample No | Origin | Sex | ESBL production | Virulence genes |
| Kp6 | urine | male | positive | ybt S,entB, mrkD |
| Kp12 | urine | female | positive | ybt S,entB, mrkD |
| Kp16 | urine | male | positive | entB, mrkD, kfu |
| Kp32 | urine | female | positive | ybt S,entB, mrkD, magA |
| Kp39 | tracheal | male | positive | entB, mrkD |
| Kp43 | urine | female | positive | ybt S,entB, mrkD, rmpA |
| Kp46 | urine | female | positive | ybt S,entB, mrkD, rmpA |
| Kp53 | urine | female | positive | ybt S,entB, mrkD |
| Kp58 | urine | female | positive | entB, iutA |
| Kp60 | urine | female | positive | entB |
Screening for ESBLs and results hypermucoviscosity K. pneumoniae (hv-KP) strains
The results of screening for the ESBL showed that 62 isolates (88.6%) were ESBL-producing . isolates. Of the 70 clinical isolates, 10 isolates (14.3%) were positive and 60 isolates (85.7%) were negative for the hv-KP test. Table 2 shows characteristics of hypermucoviscosity clinical isolates.
Virulence genes identification
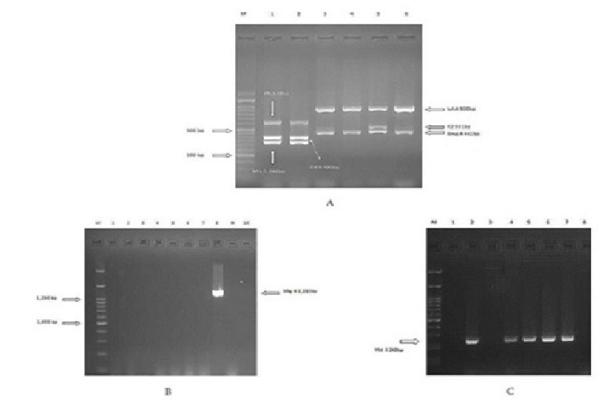
According to the results of screening virulence genes, (n= 57, 81.4%) was the most prevalent among all the clinical isolates, followed by the (n=46, 65.7 %%), (n=42, 60%), (n=8, 11.4%), (n=8, 11.4%), (n=4, 5.7%), (n=1, 1.43%), and was not detected in any of the isolates. The presence of the genes coding virulence factors was detected among the ESBL and non-ESBL isolates (

Gels electrophoresis of the PCR products in the
Frequency of virulence factor genes among the ESBL and Non -ESBL
| Total isolates | ESBL | Non-ESBL | P-Value | |
ybt S | 42(60%) | 38(90.5%) | 4(9.5%) | 0.705 |
entB | 57(81.4%) | 52(91.2%) | 5(8.8%) | 0.161 |
mrkD | 46(65.7%) | 43(93.5%) | 3(6.5%) | 0.113 |
magA | 1(1.4%) | 1(100%) | 0(0%) | 1.000 |
iutA | 8(11.4%) | 6(75%) | 2(25%) | 0.225 |
Kfu | 8(11.4%) | 7(87.5%) | 1(12.5 %) | 1.000 |
rmpA | 4(5.7%) | 4(100%) | 0(0%) | 1.000 |
K2 | 0(0 %) | 0(0%) | 0(0 %) | 0.231 |
Discussion
Generally, one of the classes of antibiotics used for treating e is beta-lactams such as cephalosporin 17. However, the presence of ESBL enzymes impairs the performance of these antibiotics 18. The difference in sensitivity and drug resistance in different geographic regions can be associated with different patterns of antibiotic use in different areas 19. In this survey, 88.6% of the clinical isolates were the ESBL producers. Moreover, as shown in the study by Ghasemi, 60% of isolates were ESBL producers in Shiraz, Iran 20. Jaskulski in Brazil reported that all isolates were ESBL-positive. The prevalence of ESBL-producing clinical isolates is related to different risk factors such as current antibiotic use, resent hospitalization 21.
has many virulence factors such as capsular polysaccharide, adhesions and siderophores which contribute to the pathogenicity of these bacteria. Presence of virulence factors in is important because they are the most prominent cause of death in patients before starting antibiotic therapy 15. genes are among genes that code virulence factors 22. Our study focused on detection of , and genes in ESBL and non-ESBL producing isolates. The important point in this study is that it was the first study to report the presence of virulencegenes in isolates in Kurdistan Province, Iran. So far, there has been no report of virulencegenes in on Google Scholar and PubMed. In the present study, was determined in 81.43% of the isolates whereas no isolates carried among all the isolates taken from the clinical specimens. Nevertheless, andwerethe highest and lowest prevalent virulence factors in the current study. In this investigation, among the 70 isolates collected from clinical specimens such as blood, tracheal, wound, and urine, 10 isolates (14.3%) were hv-KP isolates.
Frequency of was (n=4, 5.7 %) that all the isolates were ESBL. According to table 2, this is while all the hvKP-isolates had the gene and ESBL phenotype. In contrast, in previous studies, such as Yu in Taiwan, the prevalence of hv-KP, and was reported to be 38%, 48% and 17%, respectively; the result of their study showed that strains carrying were significantly associated with hv-Kp 23. On the other hand, Nahavandinejad in northern Iran demonstrated that the hv-KP isolates were not restricted to 24. was only found in one ESBL isolate that contained the and genes. In contrast, was much higher than the genes detected in Korea 25 and Taiwan 8. These difference between the prevalence of could be related to sample type of infection 26. In the majority of those studies, was isolated from liver and meninges curtains infections whereas in our study, the isolates were collected from the clinical specimens 28. In a study conducted by Feizabadi in Iran on 89 isolates of , 10 (11.2%) isolates belonged to K1 and 13 (14.6%) isolates belonged to K2 serotypes, respectively 27. Amraie in Shahrekord, Iran, reported low frequency of among clinical isolates, which is similar to our results 26. Prior studies suggested that the magA gene can be infrequently seen in isolated clinical samples except liver abscesses 2826. Compain in France designed a multiplex PCR for identifying seven virulence factors and capsular serotypes of . The multiplex PCR was used on 65 isolates between 2004 and 2014, which included 45 clinical isolates identified as hvKP; most isolates (64 /65) were found to possess 14 which is dissimilar to our results. Unfortunately, there has been no report of virulence genes in ESBL and non ESBL .
As a result of these investigations, the presence of virulence genes in ESBL-producing isolates more than clinical isolates of lacking ESBL. Our results indicated that there were no statistically significant differences between the ESBL productions and presence of the andgenes. Moreover, the presence of these genes and variables such as presence of sex, clinical specimen type and hv-KP phenotype between ESBL and non –ESBL isolates (0.05< p).
Conclusions
In conclusion, frequency of ESBL-producing is increasing now. Detection of virulence factors that positively impact the pathogenicity of is of immense importance. The results of the current study showed that was the major virulence factor for (ESBL and non-ESBL) isolated from the clinical specimens in the hospitals of Kurdistan Province, Iran.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License (CCBY4.0) which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
List of abbreviations
CDDT: Combination disk diffusion test; ESBL: Extended-spectrum betalactamase; LPS: Lipopolysaccharide; MDR: Multi-drug resistant; PCR: Polymerase Chain Reaction; UTI: Urinary tract infection
Ethics approval and consent to participate
The study was approved by Kurdistan University of medical science, Iran. All the members were fully informed of the purpose of the investigation, and were informed.
Competing interests
The authors declare no conflict of interest.
Funding
None
Authors' contributions
PS: Study design, doing experiments, data collection, writing; MKT: Study design, writing, critical review; RR: Supervision, study design, writing, critical review; AA: Data collection, data analysis, critical review; SR: Doing experiments, data collection, data analysis, critical review.
Acknowledgments
This work was retrieved from the thesis of PhD student, Pegah Shakib and, Kurdistan University of Medical Sciences supported this study.

