
Generation of Bacillus subtilis displaying alpha-toxin HlaH35LH48L fused with CotB and CotG, and studying the immune response in mice
- Center for Bioscience and Biotechnology, University of Science, Ho Chi Minh City, Viet Nam
- Vietnam National University Ho Chi Minh City, Viet Nam
- National Institute of Malaria – Parasitology – Entomology in Ho Chi Minh City, Viet Nam
Abstract
Introduction: The Bacillus subtilis spore is considered to be a powerful vehicle for surface display and antigen delivery. Being safe and widely used for the purpose of oral bacteriotherapy in humans and animals, B. subtilis spore has potential in the domain of needle-free vaccine development. The spores themselves also behave as an adjuvant. This study aims to generate B. subtilis spores expressing mutant staphylococcal alpha-toxin HlaH35LH48L on the surface and to study their ability to evoke a specific immune response in mice.
Methods: Vectors carrying the hlaH35LH48L gene fused with the coding genes of anchor proteins, cotB and cotG, were cloned in E. coli before being transformed into B. subtilis. The generation of the B. subtilis new strains via chromosomal integration was confirmed by PCR. Sporulation was observed under the microscope. The expression of the target protein on the spore surface was determined by sporeELISA. The level of IgG in the serum and IgA in the feces of Swiss mice were analyzed using ELISA to learn about the immune response against the B. subtilis spore administrated via the oral route.
Results: The PCR products of an expected size on agarose gel electrophoresis showed successful integration, resulting in the construction of new B. subtilis strains BsHT2331 and BsHT2334. sporeELISA analysis detected a significant HlaH35LH48L expression on the spore surface. The BsHT2331 spores (CotB-HlaH35LH48L) triggered a constant increase in the IgG and IgA levels in mice after three doses as 5.5-fold and 2.5-fold higher than the pre-immunization, respectively (p-value < 0.0001). Meanwhile, the group of mice orally administrated with BsHT2334 (CotG-HlaH35LH48L) showed a notable IgG titer but minor IgA response.
Conclusion: The results concluded that the construction of two new strains BsHT2331 and BsHT2334 that used spore coat proteins CotB and CotG to display mutant alpha-toxin HlaH35LH48L on the B. subtilis spore surface was successful. It provided an assessment of the ability of B. subtilis spores to stimulate specific antibody production in mice.
Introduction
is a spore-forming Gram-positive bacteria that has been developed as a major host for recombinant protein expression for decades. This is due to its highlight specialties, including (i) safety features that are commonly used in probiotic and feed additive products for humans and animals1, (ii) well-understood and easy-to-manipulate genetic system, (iii) numerous protein production capacity, and (iv) survival ability under extreme conditions2. For these reasons, spore is a potential choice for a live antigen-carrier. Not only does it remain stable in long-term storage3 but spores also have the advantage in terms of the ability to survive gastric acids and protease4, resulting in antibody responses in serum (IgG) and mucosa (fecal IgA). They have overcome the limitation of the purified protein approach in mucosal vaccine development. Besides, spores of this species have shown a significant adjuvant effect leading to the enhancement of antibody production against either co-administered antigens or spore-surface-adsorbed antigens5. These characteristics make spores an ideal bacterial vehicle to deliver antigens orally with no additional substances required.
Since the first established spore display system6, has continuously been engineered to display various pathogen antigens of viruses, bacteria, and parasites, such as Influenza A virus, and , or 1, 7. spore-based vaccines have stimulated an immune response effectively and shown there to be a significant effort to protect animal models1, 8. These properties are mainly because the endospores are formed in a multi-layered coat protein structure. Spore coat proteins are the factors that link to heterologous proteins at their C- or N-termini and anchor them to the surface. Among at least identified 80 coat proteins (7), CotB and CotG have been studied thoroughly and applied for the purpose of vaccine production9. Here we used CotB and CotG to immobilize a mutant antigen on the spore surface.
is a commensal bacterium in humans yet a pathogen once invasive, responsible for many severe diseases such as skin and soft tissue infections, bacteremia, endocarditis and pulmonary infections. The rapid increase of multidrug-resistant strains in clinical and community environments is causing difficulty in terms of treatment. The diverse virulence factors are obstacles for vaccine development. pore-forming α-hemolysin (Hla) is a common virulence that is produced in 95% of strains and it is responsible for host cell hemolytic10. Many mutation sites are created to eliminate lethality while maintaining its antigen function11. Mutation at histidine 35 has been proven to inhibit toxins from forming pores on cells, while replacing histidine 48 with leucine was needed to prevent the conversion into the natural phenotype12. It has also been studied that active immunization with an inactive variant of alpha-toxin protein, Hla, showed protection in the rabbit model12. Thus, Hlais a safe and effective candidate for antigen delivery model research. In this study, we constructed strains displaying Hla on the spore surface and learned about the immune response in murine models. Our results showed that the new strains anchoring mutant Hla were generally able to promote the IgG and IgA antibody response in animals and demonstrated the potential to develop an oral vaccine.
Bacterial strains and plasmids
|
Strains & Plasmids |
Description |
Source |
|---|---|---|
|
|
(F´ { |
Invitrogen |
|
|
|
Wu |
|
|
|
Nguyen |
|
BsHT2304 |
Control strain, containing CotB with no target gene |
Collection at Center for Bioscience and Biotechnology |
|
BsHT2305 |
Control strain, containing CotG with no target gene |
Collection at Center for Bioscience and Biotechnology |
|
pHT2331 |
|
This study |
|
pHT2334 |
|
This study |
Methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in
Primers used in this study
|
Primers |
Sequence (5’– 3’) |
Purpose |
Amplicon length (bp) |
|---|---|---|---|
|
ON2336 |
ACTGTCGCTTCCAAGACGTCGTTTGTCATTTCTTCTTTC |
|
915 |
|
ON2345 |
AGCATCAGCAGGATCCGCTGATTCTGACATCAACATCAAAAC | ||
|
ON2345 |
AGCATCAGCAGGATCCGCTGATTCTGACATCAACATCAAAAC |
Colony PCR |
943 |
|
ON1672 |
CCGGGGACGTTATTTTTCAAATTGCGGATGGCTCCAAGCAGAGACGT | ||
|
ON469 |
GGCGTTCTGTTTCTGCTTCG |
PCR for integration checking at |
1100 |
|
ON1479 |
GTCTGGTCAACTTTCCGACTCTG | ||
|
ON470 |
AACCCGCTCCGATTAAAGCTAC |
PCR for integration checking at |
1074 |
|
ON877 |
CGCTCACATTTATCGATCAATGTGATGGCTGGACAGCCTGAG | ||
|
ON2367 |
GCCGCGCGGCAGCCATATGGCTGATTCTGACATCAACATCAAAAC |
PCR for integration checking at the region between |
923 |
|
ON2368 |
GGTGGTGGTGCTCGAGTTAGACGTCGTTTGTCATTTCTTC | ||
|
ON2135 |
CGGAGCCCTGCTTATCAGCATAC |
PCR for integration checking at |
857 |
|
ON2137 |
CTGTTTGTGATGGTTATCATGCAGGATTG | ||
|
ON945 |
GCGTCCATGGAGATCTATCCGGTTGTTACTCGCTCACATTTATCG |
PCR for integration checking at |
747 |
|
ON1442 |
GATCCTCTGCCCGAAGCTCTGAC | ||
|
ON2345 |
AGCATCAGCAGGATCCGCTGATTCTGACATCAACATCAAAAC |
PCR for integration checking at the region between |
943 |
|
ON1672 |
CCGGGGACGTTATTTTTCAAATTGCGGATGGCTCCAAGCAGAGACGT |
Plasmids and strains construction
To anchor the heterologous protein on the spore surface, we used two plasmids of which design at the expression cassettes was similar to pCotB-CL and pCotG-CL9, named pHT2304 and pHT2305. The Hla gene was amplified from synthesized plasmid pHT2328 with primers ON2345 ON2336. After being cleaved with restriction enzymes I and II, the target fragment was inserted into template pHT2304 and pHT2305, which had been double-digested with the same enzymes to result in recombinant plasmids pHT2331 and pHT2334, respectively. colonies containing recombinant plasmids were selected on ampicillin LB-agar plates. The methods used to confirm the target sequences included colony PCR and sequencing.
Next, pHT2331 and pHT2334 were introduced into the strain WB80013and WB800N14, respectively. Double cross-over integration occurred during the natural transformation process to insert into the chromosome at homologous loci (for pHT2331) and (for pHT2334). colonies were selected on LB-agar plates with neomycin (for pHT2331) or chloramphenicol (for pHT2334). The correct integration of the target gene was confirmed by the PCR method with three pairs of primers per colony. All primer details are presented in
Preparation of spores
The sporulation was prepared according to the previously described method17. strains were pre-cultured in 5 mL LB medium to OD value at around 2 – 3, and then sub-cultured in 40 mL LB medium at 37C (200 rpm) to reach OD of 0.8. When the bacterial growth started entering the log phase, the cells were centrifuged at 13,000 x g for 1 minute and washed with PBS 1X. The pellet was then resuspended in 20 mL Difco Sporulation Medium (DSM) and cultivated at 37C (200 rpm) to induce sporulation. After 48 hours, the vegetative cells in the culture were eliminated by lysozyme treatment (15 mg/mL, Serva) and the spores were washed six times with PBS 1X. The spores were confirmed by Schaeffer & Fulton’s method and stored in glycerol 10% at —80C.
Schaeffer & Fulton’s method
20 µL of 48-hour cultured broth and washed spores were smeared and heat-fixed on a glass slide for 5 minutes with 5% Malachite green. The slide was cooled and washed with water to discard the dye. After that, the sample had 2.5% Safranin O applied for 30 seconds, then it was rinsed with water and dried. The results can be observed under a light microscope.
Determination of spore number
The spores' suspension at a concentration of OD value as 2 per mL was put under a heat shock at 80°C for 10 minutes to remove vegetative cells. The viable spores were counted by ten-fold serial dilution in distilled sterile water. 100 μL of each dilution was incubated on a LB agar plate overnight at 37°C. The experiment was performed in triplicate.
sporeELISA
Spores anchoring the protein on the surface, acting as an antigen, were resuspended in a 200 µL coating buffer (100 mM NaHCO; pH 9,6), and then coated onto a 96-well plate (Thermo Scientific™ Nunc™ MicroWell™ 96-Well Microplates) with a volume of 50 μL per well. After incubation at 4°C overnight, the wells were washed with PBS-Tween and blocked with a blocking buffer (PBS-Tween with 5% skim milk) for 1 hour at room temperature. After washing, 50 μL of the Hla-antibody, which was developed in Swiss mice by our research group, was added at a dilution ratio of 1/10000 incubated in 2 h at room temperature, and then washed again. The second antibody was anti-mouse IgG - peroxidase antibody produced in rabbits (whole molecule) (Sigma, A9044 – 2 mL), used at a ratio of 1/40000 at the same incubation condition. 50 μL TMB Liquid Substrate for ELISA (Sigma) was added after washing, then 50 μL HCl 1N to stop the reaction. The absorbance values were measured in a CLARIOstar plate reader at a wavelength of 450 nm. The experiment was replicated three times for each sample and presented as mean ± SD. Statistical significance was analyzed using ANOVA-one way and the Graphpad 7.0 software.
Oral immunization in mice
Each group of five 6-week-old female Swiss mice were immunized by oral gavage on days 0, 14, and 28 with 250 µL of spores BsHT2331 and BsHT2334 at a spore number corresponding to ODof 60 diluted in PBS 1X. The control groups were mice orally administrated with BsHT2304, BsHT2305, or pre-immunized mice. Serum and fecal samples were collected on day -1 (pre-immune samples), 21, and 42. Every 0.6 gram of fecal was treated with 500 uL of PBS 1X containing 0.2 mg/mL PMSF, homogenized and centrifuged at 10,000 x g in 10 minutes to collect the supernatant. All samples were stored at —20C.
Indirect ELISA
50 μL of Hla protein (5 g/mL), which was expressed in by our research group, was loaded into a 96-well plate (Thermo Scientific™ Nunc™ MicroWell™ 96-Well Microplates). The primary antibody was used as 50 µL of serum at a dilution ratio of 1/250 or fecal extract at a dilution ratio of 1/50 for overnight incubation at 4°C. Next, the plates were incubated for 2 hours with anti-mouse IgG (whole molecule) — peroxidase antibody produced in rabbits (Sigma, A9044 — 2 mL) (1/40000) or anti-mouse IgA (α-chain specific) — peroxidase antibody produced in goats (A4789 — 1 mL, Sigma) (1/10000) according to the serum or fecal sample. The IgG measurement was analyzed at room temperature, while the IgA level analysis was performed at 37°C. All samples were assayed in triplicate and read at 450 nm in a CLARIOstar plate reader. Statistical significance was analyzed using ANOVA-one way and Graphpad 7.0.
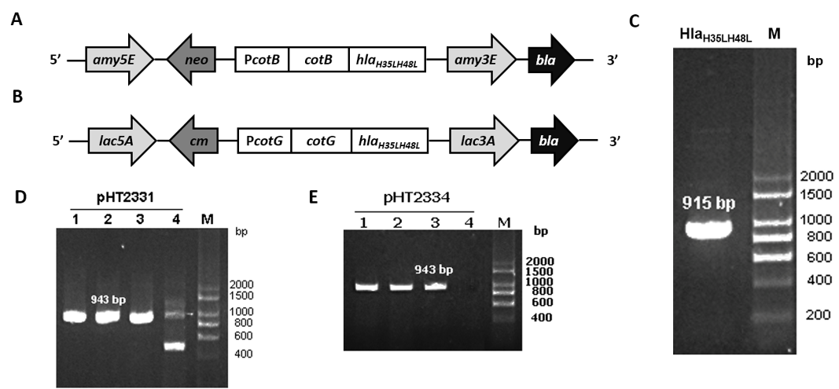
(A) Region for integration in pHT2331; (B) Region for integration inpHT2334; (C) PCR result of hla
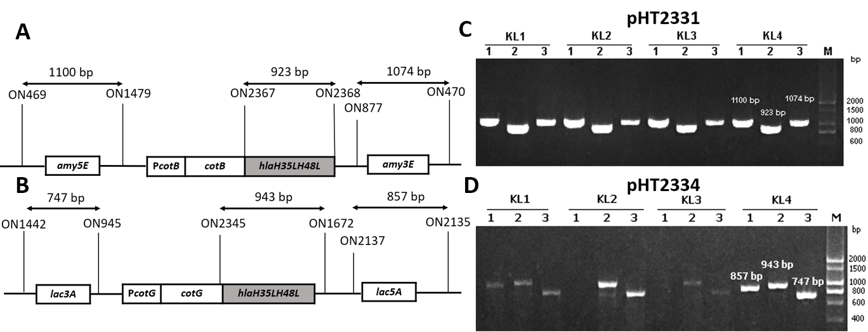
(A) Primers map for integration checking PCR of
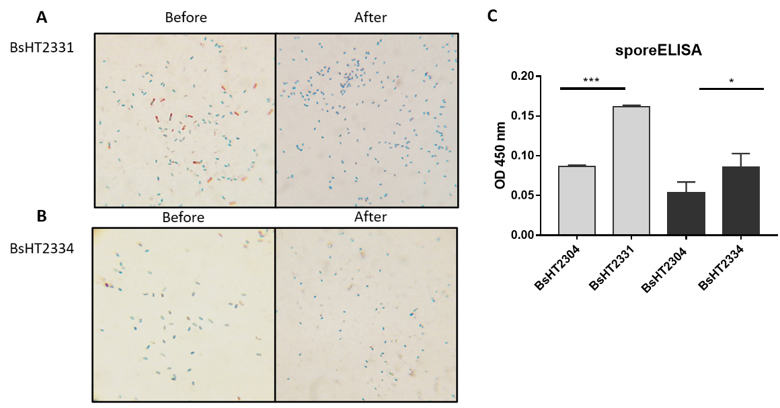
(A) BsHT2331 spore before and after lysozyme treatment; (B) BsHT2334 spore before and after lysozyme treatment; (C) sporeELISA results of BsHT2331 and BsHT2334. (ns: p-value >0.05, *: p-value ≤ 0.05, **: p-value ≤ 0.01, ***: p-value ≤ 0.001, ****:p-value ≤ 0.0001)
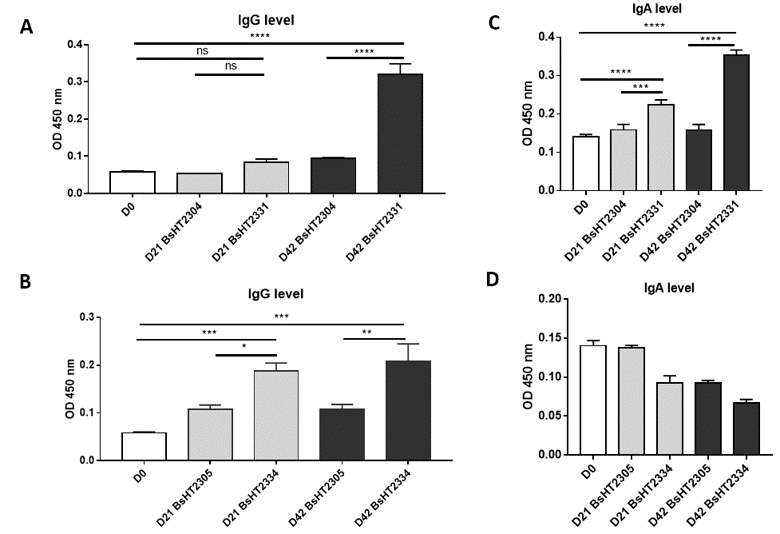
IgG and IgA level of mice oral immunized withBsHT2331 and BsHT2334 (A) IgG level of BsHT2331 on day 21 and 42; (B) IgG level of BsHT2334 on day 21 and 42; (C) IgA level of BsHT2331 on day 21 and 42; (D) IgA level of BsHT2334 on day 21 and 42. (White: Day 0; Grey: Day 21; Black: Day42) (ns: p-value > 0.05, *: p-value ≤ 0.05, **: p-value ≤ 0.01, ***: p-value≤ 0.001, ****: p-value ≤ 0.0001)
Results
Construction of vectors pHT2331 and pHT2334
The target gene was amplified through PCR using template pHT2328 with two specific primers, ON2345 and ON2336 (Figure 1C). was fused with the C-terminal region of in pHT2304 and in pHT2305 to result in two plasmid vectors pHT2331 and pHT2334, respectively (Figure 1A & B). colonies carrying the target plasmids were selected on the LB agar plate with ampicillin and by the colony PCR method which yielded the expected band of 943 bp on agarose gel (Figure 1 D & E). We confirmed via DNA sequencing that the correct sequence was inserted into the recombinant vectors (data not shown).
Generation of recombinant strains carrying integrated into the chromosomes
To generate recombinant strains carrying mutant gene, two vectors pHT2331 and pHT2334 were extracted through alkaline lysis from cells and introduced into WB800 or WB800N via natural transformation. Throughout the process, competent cells took up DNA from the environment and integrated heterologous genes into its genome at homologous loci and . These are the coding sequences of the non-essential genes of . The chromosomal integration was first confirmed via the colonies’ growth on LB agar with appropriate antibiotics. To verify that the integration of the fusion andinto chromosome occurred through a double-crossover recombination eventthree pairs of primers were used for each strain in the PCR performed. This was one pair for checking the presence of a target gene and two pairs for checking the accuracy of the integration sites, specifically and (Figure 2 A&B). The gel electrophoresis results showed that colonies with visible bands with sizes of three pairs of primers as predicted were successfully integrated strains (Figure 2 C & D). The new strains were named BsHT2331 and BsHT2334.
Display of Hla on the surface of the spores using the CotB and CotG anchor proteins
In Schaeffer-Fulton’s method, the spore coat was loosened due to the high temperature, meaning that the primary stain can penetrate the spore. The spore coat is impermeable while the cell wall has a low affinity with the water-soluble dye. Malachite green can be easily removed from vegetative cells but not the spore. Safranin is applied to re-colorize cells. In the end, the vegetative cells will color in pink and the endospores will be rod-shaped and green. Sporulation was observed under a light microscope at 100X magnification. Pictures of both BsHT2331 and BsHT2334 strains revealed an assembly consisting of pink cells, green endospores, and pink cells containing green endospores in the middle or at the end of the cell (Figure 3 A & B). This demonstrates that the sporulation reached the desired ratio and quality. The mature and free spores were then purified and their quantity determined. The density of BsHT2331 and BsHT2334 was 1.37 x 10 spores/mL and 3.83 x 10spores/mL, respectively.
To verify the presence of the target protein on the spore surface, we performed the sporeELISA experiment. Recombinant spores were considered as an antigen to interact with the polyclonal anti-Hla antibody. The spores coat was stable that allowed them to retain its original shape throughout the process. Thus, the signal released from the HRP enzyme and the substrate reaction can demonstrate the expression on the surface. The results showed that there was a significant detection of antigen Hla anchored by CotB (p-value < 0.0001) and CotG (p-value < 0.05) on the spore surface compared with each of the control strains Figure 3C). These signals reveal the presence of the target protein on the surface of the spore.
Anti-spore IgG and IgA level response by oral administration
spores can safely pass through the acidic condition of the stomach and reach the upper part of the intestine during ingestion18. Throughout the process, recombinant spores have been shown to induce both mucosal (IgA) and systemic (IgG) responses. To evaluate the antigen-delivery effect of spores displaying Hla in genetically heterogeneous animals, Swiss mice were subjected to three doses of spores on days 0, 14, and 28. Blood and fecal samples were collected on days 0, 21, and 42 for the analysis of the serum IgG and fecal IgA.
The serum IgG level of the mice orally delivered with BsHT2331 showed a slight development but not significant after the first and second doses (0.084 ± 0.009). There was a dramatic increase after the third dose (0.32 ± 0.028, p-value < 0.0001), which was 3.4-fold higher than the control strain BsHT2304 on day 42 (0.095 ± 0.001) and 5.5-fold higher than the serum sample before subjection (Figure 4A). The systemic response to BsHT2334 spore in mice also rose continuously from day 21 (0.1883 ± 0.017) to day 42 (0.2087 ± 0.036) with a 99% confidence interval compared to the mice that received BsHT2305 at the corresponding time (Figure 4B). Similarly, the mucosal immune response was determined via IgA detection in feces. Mice immunized with the BsHT2331 spore elicited a significant growth in the anti-Hlafecal IgA level on day 21 (0.2233 ± 0.014, p-value < 0.0001) compared with BsHT2305 (0.1587 ± 0.015). After three doses, the immune stimulation in mice caused by spores displaying CotB-Hla resulted in a high IgA level (0.353 ± 0.014, p-value < 0.0001) that was 2.2-fold greater than the control group BsHT2304 and 2.5-fold greater than the feces sample from day 0 (Figure 4C). Nevertheless, a moderate decrease in the IgA titer appeared from 0.093 ± 0.009 on day 21 to 0.067 ± 0.005 on day 42 in the mice that took up the CotG-Hla expression strain, BsHT2334. The ODvalue of this group also revealed no significant difference from the BsHT2305 strains (Figure 4D). The results indicate that spores expressing Hla on the surface through the CotB and CotG protein could evoke a systemic immune response in mice via the oral route. There is a difference in the IgA antibody production in the animals between the spores strains using CotB (BsHT2331) or CotG (BsHT2334) to anchor the protein.
Discussion
In this present study, we aimed to obtain recombinant spores expressing staphylococcal alpha-toxin on the surface by following a common strategy for surface display. This was built based on the application of surface molecules. Here we have genetically fused Hla to the surface-exposed spore coat proteins, CotB and CotG, and successfully integrated them into the chromosome at a homologous locus. The double cross-over integration event in is a distinctive approach to assure the genetic stability of the construct and the certain presence of the target sequences in the host cell19. During sporulation, the mutant Hla protein was under the regulation of the Cot promoter in order for it to be expressed at an accurate time for surface display20. In the sporeELISA process, is able to remain in the whole spore due to the protein on its thick coat. Thus, it was proven via the signal result of sporeELISA that both CotB and CotG have properly anchored the protein of interest on the spore coat. The antigenic feature of Hladisplaying on the spore surface was investigated and verified by the increased immunoglobulin level of the mucosal and serum responses.
Our analysis indicates that the spores using CotB showed a double high heterologous protein expression (0.1617 ± 0.002) compared to CotG (0.086 ± 0.017). In this study, we utilized the CotB sequence9 that was truncated with 105 C-terminal amino acids to enhance the efficiency of exposing the chimeric protein on the spore surface20. CotG was used in original full length. It is also noted that the genetic background of the host cell and the specific characteristics of target proteins could affect the surface expression differently. So far, CotB has been successfully applied for the purpose of vaccine development while CotG has been mainly used to immobilize enzymes on the surface21. Therefore, it is crucial to have knowledge about the most appropriate carriers among the various coat proteins for the desired antigen expression.
can cause infections in several sites in the human and animal body such as the skin and soft tissue, respiratory tract, blood stream, internal organs, and even the intestines. MRSA pneumonia cases have been reported with a high mortality rate (53 — 60%)22. Meanwhile, the intestinal carriage of is determined to be a risk factor for intestinal infection. Even though its mechanism has not been well-defined, there was histopathology evidence for a specific -induced pseudomembranous intestinal disease that was different from what was seen in usual infection by and MRSA23. Therefore, a vaccine that effectively elicits both a systemic and mucosal immune response is needed. To date, vaccine development is still a global challenge. The most common method has been the application of engineered bacteria to produce recombinant proteins or polysaccharide antigens24. There has been very little research into oral vaccination to prevent the pathogen. Thus, we aimed to develop a platform for oral vaccination using a safe and powerful tool, specifically the spore. In this research, our data revealed a choice between CotB and CotG for Hlaanchoring where CotB showed a better performance in terms of spore display and immune system stimulation. It is the fundamental information required to create further strains to express other antigens.
Conclusions
This study has successfully created two new strains, BsHT2331 and BsHT2334, that used anchor proteins CotB and CotG to display mutant alpha-toxin Hla on the sporesurface. The results of the immune response experiment following the significant increase in IgG and IgA levels provided an evaluation of the ability of spores to stimulate specific antibody production in mice.
Abbreviations
: : :ELISA: Enzyme-linked Immunosorbent assay; LB: Luria-Bertani; MRSA: Methicillin-Resistant PCR: Polymerase Chain Reaction; PMSF: Phenylmethylsulfonyl fluoride; PBS: Phosphate-buffered saline; s: TMB: 3,3′,5,5′-Tetramethylbenzidine
Acknowledgments
The author, Nhi NY Nguyen, was funded by Vingroup Joint Stock Company and supported by the Domestic Master/ PhD Scholarship Programme of Vingroup Innovation Foundation (VINIF), Vingroup Big Data Institute (VINBIGDATA) (VINIF.2020.TS.120).
Author’s contributions
HDN designed the study. NN wrote the manuscript, carried out the cloning and strains generation. HDN, LD revised the manuscript. LD, TN carried out the sporulation. NN, LD, DT, TN, AN, ThN, UN carried out the animal experiments and analyzed the data. All authors read and approved the final manuscript.
Funding
This research is funded by Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 108.06-2019.11.
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasionable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.

