Dysregulation of miR-124-3p and STAT3 in Hypothyroidism: Implications for Diagnostic and Therapeutic Biomarkers
- RNA Biology Lab, Saveetha Dental College and Hospitals, Saveetha Institute of Medical and Technical Sciences (SIMATS), Saveetha University, Chennai-77, India
Abstract
Background: Hypothyroidism is a clinical condition caused by decreased thyroid hormone (T3 and T4) levels, affecting ~5% of the population. However, its molecular mechanisms remain poorly understood. Recent studies implicate microRNAs (miRNAs) in disease-related cellular processes. This study aimed to identify novel miRNAs associated with hypothyroidism using publicly available genomic databases, with the goal of validating one candidate miRNA and its target gene in patients to evaluate their potential as diagnostic and therapeutic biomarkers.
Methods: We used bioinformatic tools (NCBI, TargetScan, miRBase) to identify candidate miRNAs, selecting miR-124-3p for further analysis. The secondary structure of miR-124-3p was predicted using RNAfold. We quantified expression levels via qPCR (quantification cycle, Cq; melt curve analysis). Similarly, we analyzed expression of Signal Transducer and Activator of Transcription 3 (STAT3), a predicted target of miR-124-3p.
Results: Secondary structure prediction revealed that miR-124-3p has a minimum free energy of -35.50 kcal/mol. miR-124-3p was downregulated and STAT3 was upregulated in hypothyroid patients, suggesting their involvement in pathogenesis.
Conclusion: Our computational and experimental data indicate that miR-124-3p is dysregulated in hypothyroidism and may serve as a diagnostic, prognostic, and therapeutic biomarker. Altered expression of miR-124-3p and its target STAT3 provides insights into hypothyroidism pathogenesis. Further studies are needed to elucidate their roles. STAT3 also shows potential as a biomarker target.
Introduction
Hypothyroidism is a chronic clinical condition characterized by reduced levels of thyroid hormones (T3 and T4), leading to elevated Thyroid Stimulating Hormone (TSH) levels1. The prevalence of subclinical hypothyroidism is comparable to that of overt hypothyroidism, affecting approximately 5% of the general population. Iodine deficiency is a major cause worldwide of chronic autoimmune thyroiditis, which accounts for the majority of cases2. Myxedema coma is a potentially fatal manifestation more prevalent among women and older individuals. Constipation, cold intolerance, fatigue, and weight gain are typical symptoms3.
The Thyroid Function Test (TFT), which measures serum TSH, T3, and T4 levels, is the standard diagnostic procedure for hypothyroidism4. Reduced T3 and T4 levels and elevated TSH levels confirm the diagnosis4. Although levothyroxine is the primary treatment, it does not cure the condition and requires lifelong adherence5. Hence, reliable biomarkers and therapeutic targets are urgently needed for improved management of hypothyroidism6.
Small non-coding RNAs known as microRNAs (miRNAs) bind to mRNAs and regulate gene expression at the post-transcriptional level7. Dysregulated miRNAs can serve as diagnostic indicators and act as tumour suppressors or oncogenes8. Notably, miR-124-3p has been implicated in various diseases, including cancer, where it can function as either a tumour suppressor or an oncogene9. Its main target is the Signal Transducer and Activator of Transcription 3 (STAT3) gene, a transcription factor crucial for numerous physiological processes, including inflammation and cell survival10.
Despite its established roles in other diseases, the role of miR-124-3p in hypothyroidism remains unexplored. Therefore, this study aims to investigate the role of miR-124-3p in hypothyroidism by examining its expression levels and those of STAT3 in patients with hypothyroidism.
Methods
Hypothyroidism-Related Gene and microRNA Sequence Retrieval
Human reference sequences for key thyroid-related genes (, and ) were retrieved from the National Center for Biotechnology Information (NCBI). These genes were selected based on their well-established roles in thyroid hormone synthesis, regulation, and action. After removing repetitive and low-quality regions, these sequences were used to construct a local nucleotide database.
Pre-miRNA and mature miRNA sequences were obtained from miRBase (http://www.mirbase.org). Candidate miRNAs were shortlisted using bioinformatic predictions (TargetScan, miRDB, miRTarBase) and prioritized based on:
-
Predicted targeting of thyroid-related genes,
-
High binding confidence scores,
-
Literature evidence supporting roles in endocrine/autoimmune pathways,
-
Biological relevance
11 .
Precursor miRNA Identification
Mature miRNA sequences were used to identify homologs in the hypothyroidism-specific database via BLAST 2.2.26+ (e-value threshold: 0.01). For non-protein-coding regions, sequences with ≤3 mismatches were retained. Putative pre-miRNA sequences within aligned regions were extracted12.
Pre-miRNA and Target Validation
Secondary structures of pre-miRNAs were predicted using RNAfold13, requiring:
-
A stable stem-loop hairpin,
-
Mature miRNA positioned on one arm,
-
≤7 mismatches between arms,
-
High A+U content and negative free energy.
Putative targets were validated using TargetScan.
Sample Collection
The study was approved by the institutional ethics committee and adhered to the Helsinki Declaration. After obtaining informed consent, blood samples were collected from 20 primary hypothyroidism patients and 20 healthy controls at Saveetha Medical College and Hospital14. Hypothyroidism was confirmed via Thyroid Function Tests (TSH, T3, T4). Samples were centrifuged and stored at –80°C.
Inclusion and Exclusion Criteria
Inclusion: Participants aged ≥18 years with informed consent.., diabetes, hypertension).
RNA Extraction and Quantification
Total RNA was extracted from blood using TRIzol reagent (Invitrogen) per manufacturer’s protocol. RNA purity and concentration were measured via NanoDrop 2000 Lite spectrophotometer (Thermo Fisher). Samples were stored at –20°C15.
Reverse Transcription
RNA was reverse-transcribed using: nuclease-free water, universal miRNA adapter, oligo(dT)₁₈ primer (Promega, 50 μM), and dNTPs (10 mM each). The mixture (10 μL) was incubated at 65°C (5 min), chilled, then expanded to 20 μL with 5× PrimeScript Buffer (NEB), murine RNase inhibitor (NEB), reverse transcriptase (NEB), and water. Cycling conditions (MiniAmp Plus Thermocycler): 30°C (10 min), 42°C (30 min), 95°C (5 min), 40°C (1 min). cDNA was stored at –20°C16.
qRT-PCR Expression Analysis
STAT3 and miR-124-3p expression was analyzed using SYBR Green (Takara) on a Bio-Rad CFX96 system. GAPDH and U6 served as endogenous controls17, 18, 19. Primers (Origene;
The primer sequences of both miR-124-3p and STAT3
|
miRNA/Gene |
Forward Sequence (5'-3') |
Reverse Sequence (5'-3') |
|---|---|---|
|
miR-124-3p |
CGCGTAAGGCACGCGGTG |
ATCCAGTGCAGGGTCCGAGG |
|
STAT3 |
GGGCATTTTTATGGCTTTCAAT |
GTTAACCCAGGCACACAGACTTC |
Statistical Analysis
Data are presented as mean ± SEM. Intergroup differences were assessed via Student’s t-test (Microsoft Excel 365), with significance at P < 0.05 (*). Pearson correlation and 95% confidence intervals for mean differences were calculated.

The secondary structure of miR-124-3p with mature sequence marked in its 3p strand.
The stem loop and mature sequence of miR-124-3p
|
No |
Structure |
Sequence (5'-3') |
|---|---|---|
|
1 |
Stem-loop |
AGGCCUCUCUCUCCGUGUUCACAGCGGACC UUGAUUUAAAUGUCCAUACAAUUAAGGCA CGCGGUGAAUGCCAAGAAUGGGGCUG |
|
2 |
Mature miRNA |
UAAGGCACGCGGUGAAUGCCAA |
The pre-miRNA length, minimum free energy, mature sequence, match extent, and A+U% content of hsa-miR-124-3p
|
Source miRNA |
Source organism |
Pre-miRNA length |
Minimum Free Energy |
Mature Sequence |
Match Extent |
Strand |
A+U% |
|
miR-124-3p |
|
85 |
- 35.30 kcal |
UAAGGCACGCGG UGAAUGCCAA |
22/22 |
3p |
49.41 |
Results
Pre-miRNA Identification and Secondary Structure Analysis
MicroRNAs (miRNAs) play a critical role in regulating gene expression and are increasingly recognized as key contributors to disease progression, including endocrine disorders such as hypothyroidism. Dysregulation of specific miRNAs could contribute to the onset or progression of this condition, highlighting their potential as early diagnostic biomarkers or therapeutic targets.
In this study, we employed a bioinformatic approach to identify candidate miRNAs associated with hypothyroidism. Reference sequences for hypothyroidism-related genes were sourced from the NCBI database, and pre-miRNA sequences were obtained from miRBase. Through sequence alignment and secondary structure prediction, miR-124-3p emerged as a candidate with strong potential relevance. Secondary structure analysis using RNAfold confirmed the mature form of this miRNA, predicting a minimum free energy of –35.50 kcal/mol, consistent with a stable hairpin structure (Figure 1;
The target genes of hsa-miR-124-3p with their molecular function and biological process
|
No |
Target Protein |
Molecular function |
Biological process |
|---|---|---|---|
|
1 |
Signal Transducer and activator of transcription 3 |
Proinflammatory gene expression |
Cell survival, cytokine release |
|
2 |
Ephrin B3 |
Regulates signalling for tissue boundaries |
Receptor tyrosine kinase |
|
3 |
Filamin B beta |
Cell structure |
Carry nutrients |
|
4 |
Tubulin gamma 1 |
Microtubule nucleating factor |
Cell division |
|
5 |
Paired box 3 |
Signalling pathways |
Embryonic development |
Target Identification
Potential targets of miR-124-3p were identified using TargetScan. The analysis revealed several significant target transcripts, including, and others (

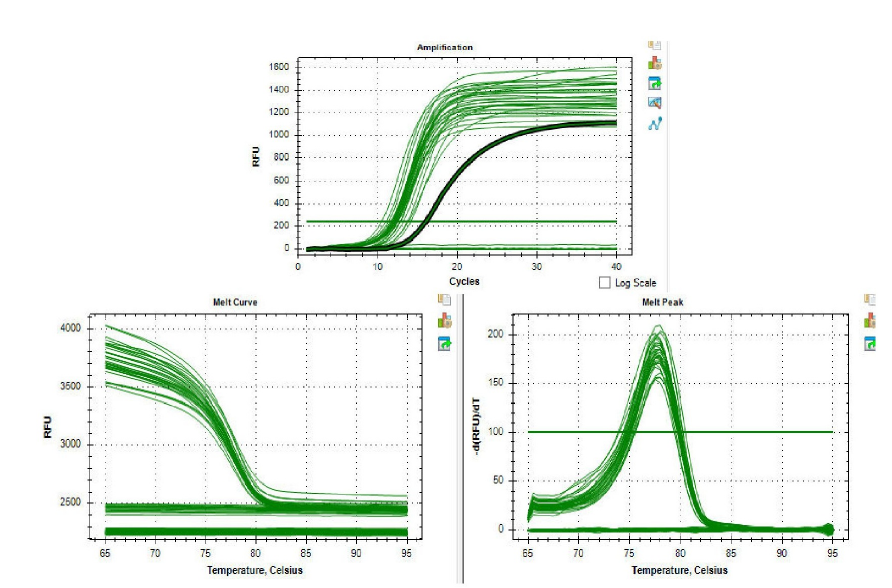
Expression Analysis of miR-124-3p in Hypothyroidism. (a) Melt curve analysis confirming the specific amplification of miR-124-3p. (b) Relative expression levels of miR-124-3p in hypothyroid patients compared to healthy controls. Data are presented as mean ± SEM. *p < 0.05, Control

Expression Analysis of STAT3 in Hypothyroidism. (a) Melt curve analysis confirming the specific amplification of STAT3. (b) Relative expression levels of STAT3 in hypothyroid patients compared to healthy controls. Data are presented as mean ± SEM. *p < 0.05, Control

The inverse relationship between STAT3 upregulation and miR-124-3p downregulation suggests a negative correlation.
Gene Expression Analysis of miR-124-3p and STAT3
We used qRT-PCR to analyze miR-124-3p and STAT3 expression in blood samples from hypothyroid patients and healthy controls. Compared with controls, hypothyroid patients exhibited significantly reduced miR-124-3p levels and elevated STAT3 levels (Figure 2 and Figure 3). The inverse relationship between STAT3 upregulation and miR-124-3p downregulation suggests a negative correlation (Figure 4), where changes in one mirror opposing changes in the other. These findings warrant further investigation into their significance in hypothyroidism.
Additionally, STAT3’s role in thyroid function remains underexplored but potentially significant. Studies indicate that thyroid hormone (T4) promotes STAT3 phosphorylation and nuclear translocation, linking thyroid hormones directly to STAT3 signaling pathways critical for thyroid function20. STAT3 also participates in autoimmune processes; its dysregulation is implicated in autoimmune thyroid diseases like Hashimoto's thyroiditis21. Hypothyroidism reduces hypothalamic and pituitary ObRb–STAT3 signaling, potentially impairing leptin-mediated thyroid regulation22. Furthermore, STAT3 may act as a tumor suppressor in thyroid cancer, highlighting its complex involvement in both normal and pathological thyroid states23. Collectively, these findings underscore the need to further explore STAT3’s role in thyroid hormone synthesis and autoimmunity in this context.
Discussion
Hypothyroidism is a common condition often caused by iodine deficiency or chronic autoimmune thyroiditis. According to recent studies, many diseases are influenced by molecular regulators, including miRNAs24, particularly miR-124-3p. Research indicates that miR-124-3p is linked to neurological conditions and malignancies. Studies show that individuals with hypothyroidism exhibit significantly lower STAT3 levels and higher miR-124-3p levels than controls, suggesting a regulatory relationship between these molecules25. By binding to the 3' UTR of target mRNAs and inducing translational inhibition or degradation, miR-124-3p typically suppresses gene expression26. However, in hypothyroidism, STAT3 upregulation may occur under specific conditions where miR-124-3p is downregulated. STAT3 is triggered by inflammatory stimuli through the JAK-STAT pathway27. The JAK-STAT system is essential for controlling cellular responses to growth stimuli and cytokines, and autoimmune disorders like hypothyroidism are associated with its dysregulation28. miR-124-3p downregulation may increase inflammation linked to hypothyroidism by exacerbating STAT3 overexpression29.
While miR-125b-5p has also been shown to target STAT3, its role differs significantly from that of miR-124-3p. miR-125b-5p is typically overexpressed in several cancers and is associated with protective effects in thyroid regulation30, 31. In contrast, miR-124-3p appears to have a more disease-specific role in hypothyroidism, where its overexpression correlates with STAT3 suppression. This distinction suggests that miR-124-3p is more specifically involved in autoimmune-related thyroid dysfunction, while miR-125b-5p may exert broader anti-inflammatory or oncogenic effects. Unlike miR-125b-5p, which is associated with generalized suppression of STAT3 in multiple cell types, miR-124-3p may serve as a more context-specific biomarker or therapeutic target in hypothyroidism.
Although our study used TargetScan to predict STAT3 as a potential target of miR-124-3p, this interaction has already been experimentally validated in previous research. Luciferase reporter assays have confirmed that miR-124-3p directly binds to the 3' UTR of STAT3, leading to its post-transcriptional repression32, 33. Thus, our findings are supported by established functional evidence, strengthening the biological relevance of the miR-124-3p/STAT3 regulatory axis in hypothyroidism. Importantly, cellular context influences the miR-124-3p/STAT3 relationship. Though expression data support direct regulation, indirect effects are possible. Future studies should explore the interplay of miR-124-3p with other regulatory mechanisms.
To normalize the expression of STAT3 and miR-124-3p in RNA isolated from peripheral blood, we used GAPDH and U6 snRNA as reference genes. Although the stability of these reference genes in whole blood has been questioned, our procedure—including whole RNA extraction and cDNA synthesis—minimizes the direct impact of blood matrix variability. Both GAPDH and U6 are frequently employed as internal controls in gene and miRNA expression studies using blood-derived RNA due to extensive validation34, 35. Additionally, to preserve RNA integrity and minimize technical variability, all samples were processed under uniform experimental conditions. However, we acknowledge that reference gene expression can vary with disease stage. Thus, future research should incorporate additional reference genes or alternative normalization techniques to verify expression stability.
A major limitation of our study is its small sample size (20 patients and 20 controls). Though expression differences in STAT3 and miR-124-3p reached statistical significance, the small cohort size may limit the robustness and generalizability of our findings. No formal power analysis was performed during study design. To confirm and expand these findings, future studies must include larger, more diverse populations and appropriate power estimates.
Finally, correlating miR-124-3p and STAT3 levels with clinical thyroid parameters (TSH, T3, T4) is essential for substantiating their utility as biomarkers. We hypothesize a positive correlation between miR-124-3p and TSH and a negative correlation with T3/T436. Conversely, STAT3 expression is expected to positively correlate with T3/T4 and negatively with TSH37. These associations would strengthen the clinical relevance of miR-124-3p and STAT3 as thyroid function indicators.
Although our study provides insight into miR-124-3p and STAT3 expression, its narrow focus limits exploration of broader roles or interactions with other hypothyroidism-related genes. Future research must clarify the precise molecular mechanisms governing miR-124-3p and STAT3, including feedback loops and cross-talk with other signaling pathways. Confirmatory studies in larger, diverse cohorts are needed to investigate connections between miR-124-3p, STAT3, and other disease processes. We propose miR-124-3p as a potential therapeutic target and diagnostic biomarker for hypothyroidism. Nevertheless, further research is necessary to fully understand its role and translational applications.
Conclusions
Our findings suggest that miR-124-3p is downregulated in hypothyroidism, while its target STAT3 is upregulated, indicating a critical role for this regulatory axis in thyroid function and disease pathogenesis. Notably, miR-124-3p’s dysregulation supports its potential as a diagnostic, prognostic, and therapeutic biomarker. Likewise, elevated STAT3 expression underscores its mechanistic involvement in hypothyroidism and its promise as an additional therapeutic target. Further large-scale, multi-center studies are warranted to validate these observations and to clarify miR-124-3p/STAT3 interactions in diverse patient populations, ultimately guiding the development of miRNA-based interventions to improve hypothyroidism management.
Abbreviations
BLAST – Basic Local Alignment Search Tool, cDNA – Complementary DNA, cT – Cycle Threshold, dNTP – Deoxyribonucleotide Triphosphates, GAPDH – Glyceraldehyde-3-Phosphate Dehydrogenase, JAK-STAT – Janus Kinase-Signal Transducer and Activator of Transcription, miRNA – microRNA, mRNA – Messenger RNA, NCBI – National Center for Biotechnology Information, ncRNA – Non-coding RNA, OSCC – Oral Squamous Cell Carcinoma, PCR – Polymerase Chain Reaction, qRT-PCR – Quantitative Reverse Transcription Polymerase Chain Reaction, RNA – Ribonucleic Acid, SEM – Standard Error of the Mean, STAT3 – Signal Transducer and Activator of Transcription 3, T3 – Triiodothyronine, T4 – Thyroxine, TFT – Thyroid Function Test, TSH – Thyroid Stimulating Hormone, UTR – Untranslated Region, U6 – U6 Small Nuclear RNA (snRNA).
Acknowledgments
None.
Author’s contributions
Original draft preparation: A.K; Formal analysis: D.S; Editing and analysis: P.K; writing – review and editing: A.P. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by an Institutional Review Board (IRB), Saveetha Dental
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.

